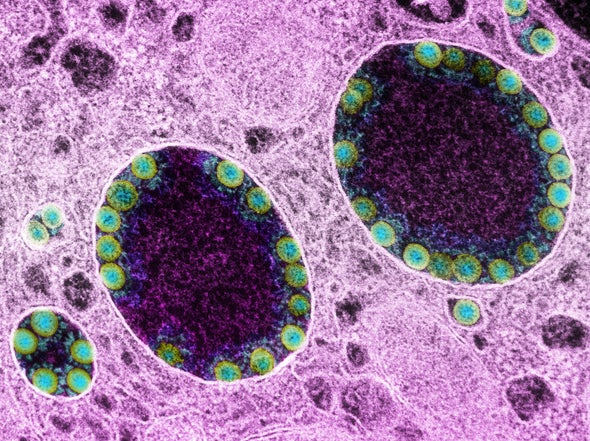
Virologist Sissy Sonnleitner tracks nearly every COVID-19 case in Austria’s rugged eastern Tyrol region. So, when one woman there kept testing positive for months on end, Sonnleitner was determined to work out what was going on.
Before becoming infected with SARS-CoV-2 in late 2020, the woman, who was in her 60s, had been taking immune-suppressing drugs to treat a lymphoma relapse. The COVID-19 infection lingered for more than seven months, causing relatively mild symptoms, including fatigue and a cough.
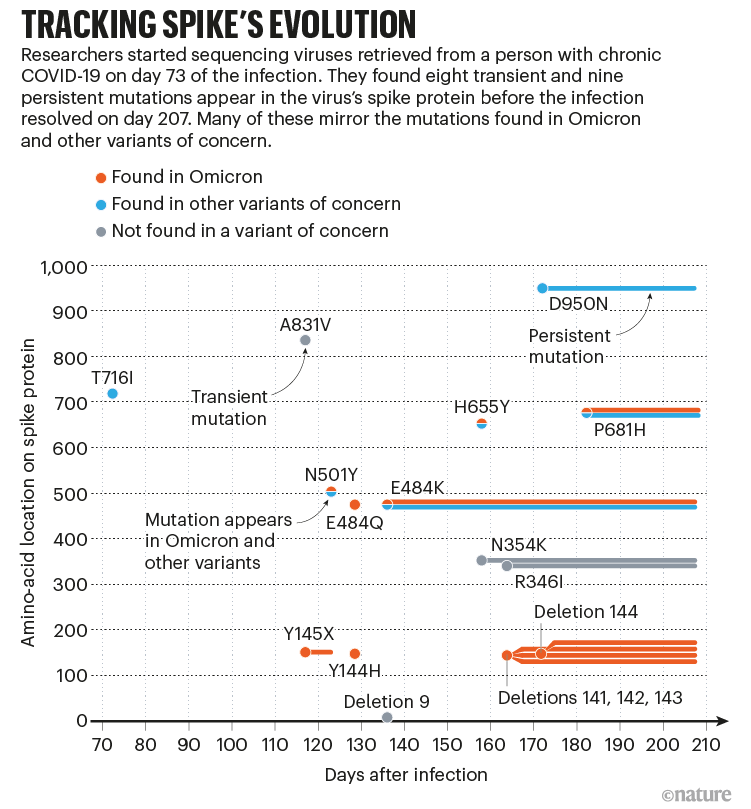
Sonnleitner, who is based at a microbiology facility in Außervillgraten, Austria, and her colleagues collected more than two dozen viral samples from the woman over time and found through genetic sequencing that it had picked up about 22 mutations (see ‘Tracking spike’s evolution’). Roughly half of them would be seen again in the heavily mutated Omicron variants of SARS-CoV-2 that surged around the globe months later. “When Omicron was found, we had a great moment of surprise,” Sonnleitner says. “We already had those mutations in our variant.”
Omicron did not arise from the woman’s infection, which doesn’t seem to have spread to anyone. And although no definitive links have been made to individual cases, chronic infections such as hers are a leading candidate for the origins of Omicron and other variants that have driven COVID-19 surges globally. “I don’t think there can be any doubt in anyone’s mind that these are a source of new variants,” says Ravindra Gupta, a virologist at the University of Cambridge, UK.
Researchers want to understand how the virus might evolve the ability to spread from person to person more easily, to evade the immune response, or to become more or less severe. Some or all of these qualities might be forged during the course of a chronic infection. “We don’t quite understand what can evolve in a single individual — and what cannot,” says Alex Sigal, a virologist at the Africa Health Research Institute in Durban, South Africa.
The odds are remote that this knowledge could help to predict the next deadly strain or even to trace variants such as Omicron to their origin. Still, virologists hope that by improving their understanding of viral evolution, they will be able to anticipate what future variants might look like — and potentially find better ways to treat chronic infections. “It’s such an important problem, given that we don’t want another variant that we can’t handle,” says Sigal.
Deadly competition
Since late 2019, scientists have sequenced the genomes of more than 11 million samples of SARS-CoV-2 taken from people. These efforts have drawn an evolutionary tree that is remarkable in its breadth, showing how the virus has changed during its march around the planet, gaining just a couple of stable mutations per month as it moves from person to person.
“But that’s only one part of the evolutionary story,” says Sarah Otto, an evolutionary biologist at the University of British Columbia in Vancouver, Canada. Each person’s infection is its own universe, where new mutations arise as the infection spreads from cell to cell. Most of these changes won’t matter to the virus, and many will do it harm. But some might give it a slight advantage over other versions of the virus in that person’s body, enhancing its ability to spread or providing some resistance to immune defences. These two traits — infectivity and immune evasion — are the main ways in which SARS-CoV-2 has evolved since it first emerged in 2019.
In acute SARS-CoV-2 infections, which generally last a week or two before being cleared by the immune system, versions of the virus with advantageous mutations have little time to outcompete those that lack them. The odds of a virus with such an advantage being transmitted to another individual are therefore small. Studies suggest that only a few virus particles — maybe even just one — are needed to seed a new infection. “Which of those viruses happens to be in the aerosol droplet you sneeze out at the time someone walks by and breathes in is largely a matter of luck,” says Jesse Bloom, a evolutionary biologist at the Fred Hutchinson Cancer Center in Seattle, Washington. “So, most of the beneficial mutations that have arisen in a patient are lost, and then evolution has to start up all over again.”
This ‘transmission bottleneck’ is the reason SARS-CoV-2 picks up around two mutations per month globally, on average. But in chronic infections, which last for weeks to months, viruses with advantageous mutations have time to outcompete others.
Compared with acute cases, these long-term infections also allow time for much more viral diversity to develop. And through a process called recombination, which can shuffle the genomes of SARS-CoV-2 particles together, mutations that are beneficial in one part of the body, such as the upper airways, might show up in viruses bearing other useful properties, says Andrew Rambaut, an evolutionary biologist at the University of Edinburgh, UK. “If the result is a fitter virus, it can suddenly take off.”
As a result of chronic infections, globally, “this virus has opportunities not just to evolve in one way, in one direction, but literally thousands, maybe tens of thousands of directions over months”, Otto says.
Targeting spike
No two chronic infections are identical. But in dozens of case reports, researchers have begun to identify common signatures of long-term infection. One of the most striking, says Otto, is the large number of amino-acid changes that accrue in the virus’s spike protein, which helps it to infect cells and is a primary target for the body’s immune response.
Many of these mutations map to regions of the spike that are targeted by antibodies, such as its receptor binding domain (RBD) and the N-terminal domain, which are involved in recognizing and infecting host cells. This makes sense, says Darren Martin, an evolutionary virologist at the University of Cape Town in South Africa. If a person’s immune system fails to clear an infection fully, the surviving viruses are likely to bear immunity-evading mutations that helped them to survive the attack. One study, which has not been peer reviewed, found that the most common mutation in chronic infections is at a position in the spike protein’s RBD called E484. Changes at this site can prevent some potent infection-blocking antibodies from attaching to the virus.
Some mutations don’t work particularly well on their own. Last year, Gupta and his team described a 102-day infection in a man in his 70s who had a compromised immune system, and who ultimately died from the infection4. After doctors had treated him with convalescent plasma — the antibody-containing portion of blood donated by people who had recovered from COVID-19 — Gupta’s team found that viruses with a pair of spike-protein mutations were thriving in the man’s airways.
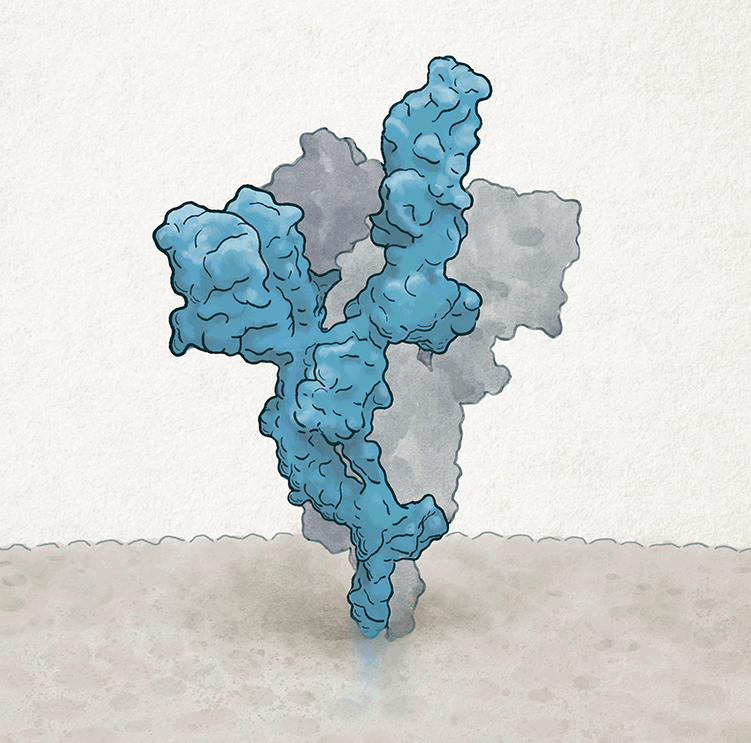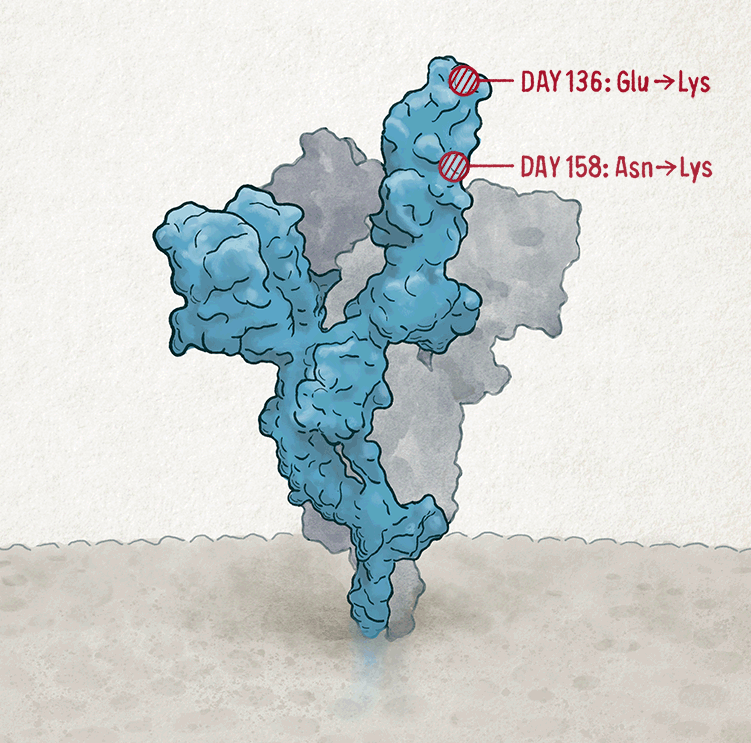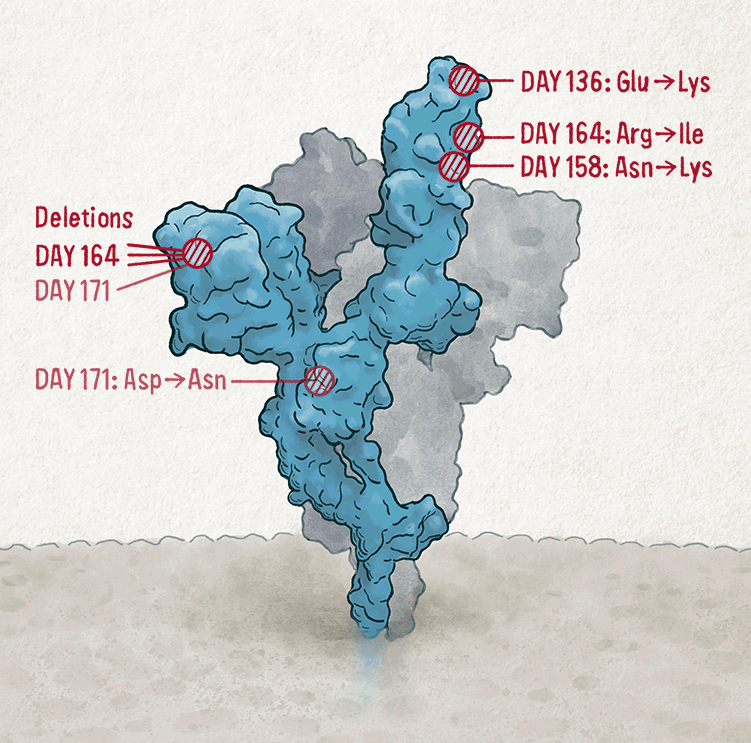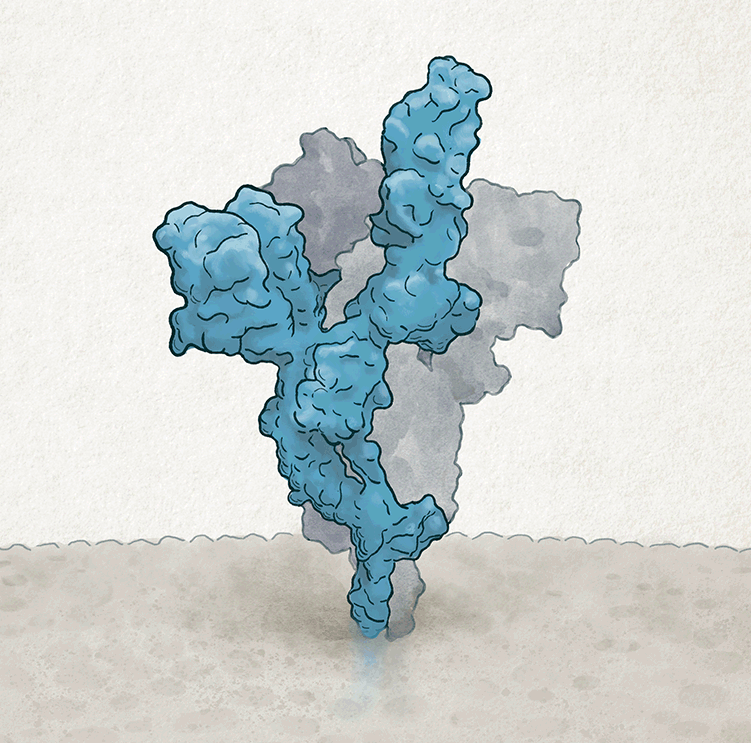
One of the mutations, called D796H, conferred resistance to antibodies — but this benefit came at a cost to the virus. When the researchers engineered a non-replicating ‘pseudotype virus’ to carry the D796H mutation and measured how well it could infect cells in the lab, they found that this mutation alone made the pseudotype virus significantly less infectious. But when the pseudotype virus also contained a second mutation found in the same person — a two-amino-acid deletion at sites 69 and 70 — infectivity was restored almost completely. Such compensatory mutations, which have more time to emerge in chronic infections, allow the virus to make evolutionary leaps, says Gupta. “Viruses struggle to do that when they’re jumping between hosts very quickly.”
In some cases, mutations have made sense only with hindsight. In late 2020, Jonathan Li, a physician-scientist at Brigham and Women’s Hospital in Boston, Massachusetts, and his colleagues released the first detailed report of a chronic SARS-CoV-2 infection: an ultimately fatal case in a 45-year-old man who had a rare autoimmune disease. The virus developed mutations linked to antibody resistance, including E484K, and another spike mutation called N501Y, which lab studies had suggested improves the virus’s ability to bind to host-cell receptors, potentially boosting infectivity.
The significance of the N501Y change became apparent when it was detected in a trio of fast-growing lineages later named the Alpha, Beta and Gamma variants of concern (VOCs). Omicron bears this mutation, as well as several others identified in the man’s infection. “He really was the harbinger of what was to come,” Li says.
Seeking variant origins
Alpha, identified in the United Kingdom in late 2020, was the first SARS-CoV-2 variant suspected to have emerged from a chronic infection. But that wasn’t the only possible explanation, says Rambaut. The variant might have arisen in a region — probably outside the United Kingdom — that had little capability to conduct genomic surveillance of SARS-CoV-2. Alternatively, Alpha could have evolved in an animal reservoir (the variant’s N501Y mutation enables it to infect mice, rats and mink).
A chance discovery nevertheless suggests that a chronic infection was the most likely source of Alpha. Rambaut and Verity Hill, an evolutionary biologist at the University of Edinburgh, reported in a March preprint the discovery of an intermediate version of Alpha in UK sequencing data. The sequence was collected from a person in southeast England in July 2020, two months before Alpha was first detected in the same region.
The virus had acquired the N501Y mutation, as well as several other hallmarks of Alpha, but it lacked the full suite of changes. “It’s accumulating these mutations. It was probably a bit rubbish at spreading,” Hill says. Only once the Alpha intermediate gained further mutations did it have the capacity to take off, she suggests.
Combinations of mutations are seen in Omicron, too. That variant — which includes several sub-lineages with many overlapping mutations — is brimming with genetic changes linked to both immune escape and infectivity that had been spotted before. But what stood out to Martin was that the BA.1 subvariant that set off most countries’ Omicron waves has a collection of 13 spike mutations that scientists had rarely seen individually, let alone all together in a single virus.
Martin and his colleagues hypothesize that, among this unique set of mutations, are some that helped to offset the evolutionary costs associated with the mutations that hastened Omicron’s spread. “Those trade-offs take a long time to resolve and those require, in my opinion, chronic infections,” says Martin. These could be in humans or in animals, he adds.
Another characteristic of Omicron — the reduced severity of disease — could also be a product of chronic infection. Lab studies have suggested that Omicron’s relative mildness could be a result of its preference for infecting cells in the upper airways, as opposed to those in the lung. The variant probably evolved from a strain that adeptly infected both upper and lower airways. Gupta suspects that Omicron’s shift probably depended on the kind of coordinated evolution that occurs when a virus spends months in a single person’s body. But what’s not clear are the evolutionary forces that propelled such a shift, he adds.
On the lookout
Chronic infections could be the best explanation for how variants such as Omicron and Alpha evolved. But it’s not obvious how one of the defining characteristics of most variants — their ability to spread like wildfire between people — might evolve in a single individual. “That’s a real mystery,” says Bloom. “When something’s not under selection, you often lose it. During a chronic infection there’s no longer selection for transmissibility.”
One possible explanation is that the same molecular mechanisms that help SARS-CoV-2 to infect a person’s airways, lungs and other organs are also important for enabling the virus to spread to others. “The same transmission dynamics are required when it’s inside you as when it’s going from one person to another,” says Martin.
But there is a difference between a virus that merely retains the ability to transmit, and one such as Omicron or Alpha that can cause a global surge in cases. A massive boost in transmissibility or the capacity to infect previously immune people might be what sets a dangerous VOC apart, says Rambaut. “It’s not that all chronic infections are going to produce VOCs. It’s going to be one in a million.”

That means that surveillance is unlikely to detect a variant at its point of emergence. In a May preprint, researchers spotted an Omicron strain that had picked up other spike mutations during chronic infection in an immunocompromised individual, and showed that it had spread to several people in the same hospital, as well as in the local community. But wider spread of such infections seems exceedingly rare. A February preprint documenting 27 people with chronic infections reports no evidence that any had spread the virus to other individuals. If VOCs so rarely emerge from chronic infections, it will be difficult to prevent them without reducing overall rates of infection around the world, says Adi Stern, an evolutionary virologist at Tel Aviv University in Israel, who led the study.
Nevertheless, there is an urgent need to understand the viral factors that contribute to chronic infections. “We need to go beyond the case reports and understand what the virus is actually evolving during this time,” says Sigal.
Sigal and his team are tracking people with advanced HIV, whose immune systems can be severely compromised, to identify factors associated with chronic SARS-CoV-2 infection. HIV infects immune cells called CD4+ T cells, which also support the production of antibodies against viruses such as SARS-CoV-2. In unpublished work, Sigal and his colleagues have found that low levels of CD4+ T cells are associated with a risk of chronic SARS-CoV-2 infection, and that many of the cases are mild, with few or no respiratory symptoms.
On the basis of the sheer number of people living with HIV — nearly 40 million globally — and the likelihood that most people have already been infected with SARS-CoV-2, it seems likely that some cases of persistent infection are contributing to the emergence of new variants, says Otto. “From an Occam’s razor point of view, we know that should be a source.”
People with compromised immune systems aren’t the only potential source of variants. Researchers have documented SARS-CoV-2 infections lasting multiple weeks in people with healthy immune systems. From the perspective of natural selection, even a relatively short three-week infection provides exponentially more opportunities for the virus to evolve, compared with an acute infection lasting a week, says Martin.
People with relatively healthy immune systems might also provide the virus with more selection pressure than individuals who have impaired immune responses, says Hill. But how to identify people who are susceptible to such infections or what their symptoms might look like is an open question. “I would suspect they’re a lot more common than we realize,” says Hill.
Last year, Gonzalo Bello, a virologist at the Oswaldo Cruz Institute in Rio de Janeiro, Brazil, and his colleagues identified several strains of SARS-CoV-2 circulating in Amazonas state in Brazil. These carried some — but not all — of the mutations found in the Gamma variant that drove the region’s ferocious second wave in 2021. But each of the Gamma-like strains also had their own unique mutations: evidence, Bello says, that Gamma might have evolved not from a single chronic infection, but from transmission chains of medium-length infections involving relatively healthy people.
Such transmission chains could have contributed to the diversity of Omicron lineages, Bello suggests. “Maybe these individuals are where some of the steps in the origin of VOCs are happening,” he says. And if chronic infections in healthy people are a likely source of VOCs, improving global vaccination rates could help to prevent new ones emerging, Hill adds. “When you’ve got these huge uncontrolled waves of infection, you’re sowing the seeds for the next.”
Antiviral drugs and other treatments taken during a chronic infection could also be playing a part in the virus’s evolution. One trait scientists are looking out for is resistance to COVID-19 drugs such as Paxlovid (nirmatrelvir–ritonavir) and molnupiravir. (Resistance to the antiviral remdesivir has already been documented in chronic infections.) The drugs affect highly conserved viral proteins — for which the barrier to drug resistance is high — but evolutionary leaps that characterize chronic infections could buy the virus time to come up with a way around that, says Gupta.
In unpublished laboratory experiments, a team led by virologist David Ho at Columbia University in New York City has found that SARS-CoV-2 can take numerous paths to Paxlovid resistance. Some involve gaining compensatory mutations that allow the virus to overcome the costs of Paxlovid resistance, allowing them to thrive, at least in the lab. Such mutations are unlikely to be behind anecdotal reports of recurring SARS-CoV-2 symptoms after Paxlovid treatment, says Ho (who himself experienced such a rebound). But if the treatment, which is normally taken for five days, is administered for a longer period to treat a chronic infection, there is a good chance resistance will emerge.
There is also an urgent need to identify effective treatments for chronic infections — particularly in people with immune-system impairments, who don’t always mount a strong response to vaccines. Most approved monoclonal antibody drugs are not effective against Omicron and its offshoots, and researchers have shown in a preprint that resistance to these therapies can emerge when they’re used to treat chronic infections.
Convalescent plasma should create a higher evolutionary barrier than monoclonal antibody therapies, says Arturo Casadevall, a microbiologist at John Hopkins Bloomberg School of Public Health in Baltimore, Maryland. Plasma that contains high levels of diverse antibodies has been shown to be effective at treating COVID-19, and some physicians are now giving it to people with compromised immune systems.
Antiretroviral drugs that target HIV can also help people living with that virus to clear chronic SARS-CoV-2 infections, but adherence to the drugs can be a challenge, Sigal notes.
Last October, UK clinicians reported a case in which a person’s chronic infection was cleared after they received a COVID-19 vaccine16. For the Austrian woman whom Sonnleitner and her colleagues studied, the end of her seven-month infection also followed vaccination. But it’s impossible to know if the vaccine is what helped her to recover.
That outcome is rare for people with chronic infections, however; many reports end in death. “They really are heartbreaking cases,” Stern says. As many parts of the world attempt to move on from the pandemic, with some healthy people shrugging their shoulders at ‘mild’ Omicron infections, Stern says we must do more to protect those who are most at risk of a chronic SARS-CoV-2 infection. “It’s dangerous for them — and it’s dangerous for us as a society.”
This article is reproduced with permission and was first published on June 15 2022.