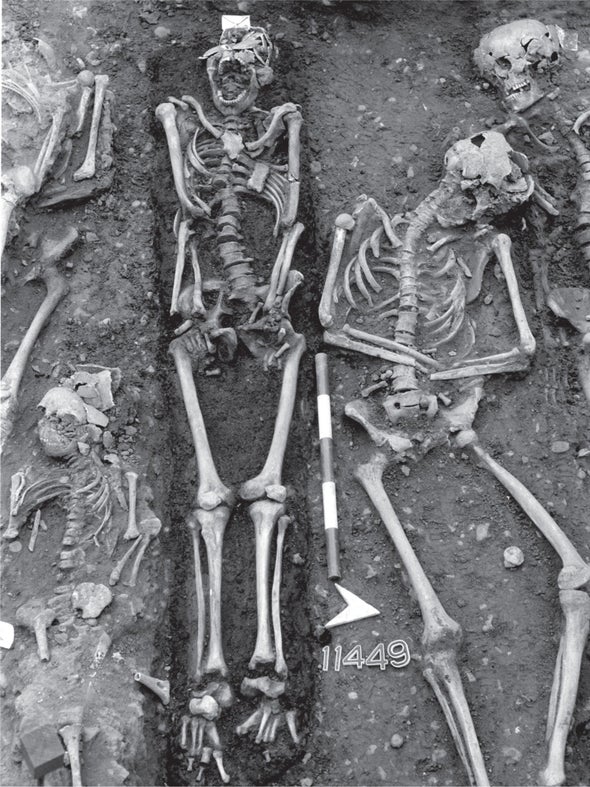
In 541 C.E., after years of campaigning against Goths and Vandals, Emperor Justinian I had built the eastern Roman Empire into a vast dominion, nearly encircling the Mediterranean Sea. That year, however, gave the ruler no chance to celebrate. Instead he was attacked by a deadly new foe, an invisible and incomprehensible enemy.
A mysterious plague swept across Justinian's lands and into his capital, Constantinople. Victims spiked high fevers, their armpits and groins swelled painfully, and many became delirious. The emperor himself fell ill. Rumors of his demise filled the panicked city. Historian Procopius, a resident of the city, claimed that on some days as many as 10,000 people died. Justinian managed to survive the contagion, but his empire remained scarred for years afterward, losing its grip on many territories and struggling to maintain control of Rome.
Scientists have debated the identity of this scourge up to the present day. While some blamed the plague on a particularly lethal strain of the bacterium Yersinia pestis—the symptoms resembled the medieval Black Death, and Y. pestis is the bug behind that devastation—other have argued Justinian was beset by an influenza virus related to the notorious 1918 flu epidemic, which killed an estimated 50 million to 100 million people. Historians have also wondered where the disease started. Many pointed the finger at Egypt because historical accounts noted a similar ailment appeared there just before Justinian's catastrophe.
Now biologists and archaeologists, teaming up to recover ancient DNA from teeth and bones from that time, have been able to resolve this long-standing debate. The teeth hold DNA from Y. pestis, not remnants of the flu. Following this strain back in time and across the globe, researchers learned that the plague began not in Egypt but in western China and traveled across the high grasslands of the Eurasian Steppe before hitting Europe.
The disease “had evolved for quite a while before it was seen in the Roman Empire,” says Alexander Herbig of the Max Planck Institute for the Science of Human History in Jena, Germany, who used computers to reconstruct DNA changes in the pathogen as it moved from place to place. Over time some of these changes enabled the pathogen to live and spread in new kinds of hosts, extending its devastating reach.
This ability to pull the DNA of disease-causing microorganisms from ancient human remains is helping to fill in a number of big blanks in history books. The molecules show how our history has been shaped by encounters with bacteria and viruses that blew up into pandemics. In addition to the events that shook Justinian's era, scientists have used pathogen DNA to improve our understanding of the origins of the Black Death and the fall of the Aztec Empire. They have even found evidence that a disease during the Bronze Age paved the way for a surge of people out of Asia and into Europe—and those people brought with them technology, culture and genes whose influence can still be seen today.
With these discoveries, certain patterns have emerged about the way microbes turn into plagues. The tiny organisms tend to spread death when they encounter groups of individuals who live packed densely together. They race through populations that have never been exposed before and thus have low levels of natural immunity. Growing international trade and increased human mobility amplify the spread, and pathogens usually have found heightened vulnerabilities among people marginalized and impoverished by society, who have few resources to protect themselves. We are now seeing these patterns again as our current pandemic, driven by the SARS-CoV-2 virus, races across the globe.
Molecular Clues
Scientists and historians have long been interested in connecting pathogen biology to history, but until about a decade ago attempts were thwarted by difficulties in analyzing DNA from ancient remains (aDNA). Efforts to recover pathogen genomes from Black Death burials, for instance, resulted only in “repeated failure, repeated failure, repeated failure,” laments Hendrik Poinar, an aDNA biologist at McMaster University in Ontario. The buried bones were degraded and held only vanishingly small quantities of genetic molecules belonging to the microbes.
Two developments changed the picture in the past decade. One was the recognition by specialists in archaeological genomics that they had been looking at the wrong parts of the skeleton. Teeth, not bones, are the best time capsules. On the outside, teeth are shielded by a tough enamel layer. On the inside, the dental pulp is stuffed with desiccated blood—and the degraded remnants of blood-borne pathogens. Scientists can drill out the interior with a dental drill, dissolve it and, with a little luck, discover some remnants of microbial DNA.
But those samples are scrambled and fragmented. They still need to be stitched back together into long, detailed sequences of DNA so that they can be identified as belonging to a particular bacterium or virus. Next-generation sequencing, a method that speeds up this reassembly, was the second big advance. The technique came into wide use recently thanks to more powerful computers, and it “changed the game entirely,” Poinar says. Essentially the method involves sequencing lots of short strings of DNA at the same time, in parallel, and reassembling them into a recognizable genome by connecting them where series of letters (the familiar A, T, C and G of the genetic code) overlap. The approach makes it possible to reconstruct an entire genome from a degraded sample, sidestepping the need to recover a rare long stretch of high-quality DNA.
One of the first successes of this combination of better samples and new technology came in 2011. Poinar and his colleagues recovered a draft Y. pestis genome from teeth obtained in a London Black Death burial site. Their find confirmed, after decades of speculation, that this bug was indeed responsible for the medieval pandemic that killed 30 percent or more of the European population between 1347 and 1351. There was nothing especially virulent about this strain, researchers learned over the next five years; it was quite similar to modern Y. pestis, which is not nearly as deadly. The high medieval death toll seemed to be driven by an exploding population of runaway black rats, which carried the bacterium through a crowded and malnourished population in burgeoning cities with awful sanitary conditions.
Perhaps the biggest surprise from plague aDNA has come from even earlier burials. It turns out that neither the Justinian nor the medieval pandemics were the first times this microbe altered human events on a transcontinental scale.
A Prehistoric Pandemic
In 2015 data from 101 ancient human genomes, extracted from skeletons buried across the Eurasian Steppe, established that an Early Bronze Age people, the Yamnaya culture, moved down from the steppe around 5,000 years ago, replacing the Neolithic farming cultures of Europe. The newcomers had domesticated horses and new forms of metallurgy and were probably warlike, but still the large-scale population changeover has puzzled scientists because the European groups had done well for centuries. “How on earth could those very well-organized, apparently prosperous European Neolithic societies go into decline?” asked one of the archaeologists involved in this work, Kristian Kristiansen of the University of Gothenburg in Sweden. Some archaeological evidence pointed toward a Europe-wide population crash among the farmers around the time of the Yamnaya arrival, and Kristiansen says he and his colleagues started to wonder if a disease had weakened European society enough to allow for an incursion. In particular, he asked: “Could it be the Black Death?”
When Kristiansen's team had sequenced the 101 ancient steppe human genomes, they had scooped up everything—not just human DNA but a soup of viruses, bacteria and modern environmental contamination. “Between 95 and 99 percent of the data we didn't use,” says team member Simon Rasmussen of the University of Copenhagen. “We were just throwing it into the dumpster.” But by 2015 the new sequencing technology had given them the ability to sift through the material and compare it with other genomes. “So we took all these data—100 billion small pieces of DNA—and we screened them against the plague,” Rasmussen says.
In about two weeks the scientists’ machines came up with an answer. Around 7 percent of the ancient remains had traces of plague DNA lingering inside their teeth. Bacteria get into teeth via blood vessels, and “if the bacteria are in the blood, that's really bad,” according to Rasmussen. “Very likely these people died from it.” This more lethal form of the disease is called septicemic plague; in the most common type, bubonic plague, the bacterium infects the lymph nodes. Because many aDNA samples they examined were in poor shape, Rasmussen also suspects that the plague DNA might be present in an even higher proportion of the material but was too scrambled to detect. He thinks the evidence is “starting to edge toward a likely pandemic.” Poinar, however, is more cautious and notes that other factors besides Y. pestis—such as famine or warfare—could have contributed to the European population crash.
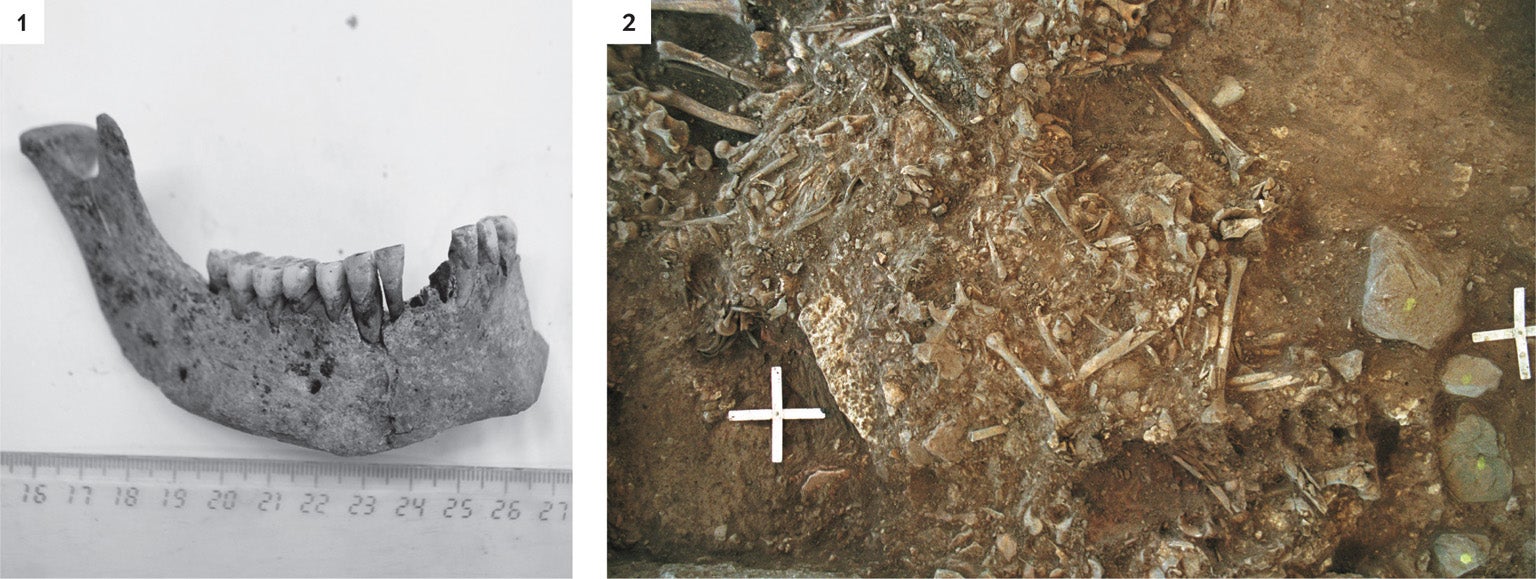
If the plague bacterium was even part of the cause, the effects can be seen today. Kristiansen's team argues that, just like the later Y. pestis outbreaks, this one spread from the steppe into Europe. The Yamnaya likely had some immunity to the bacterium if they had already been exposed to it for hundreds of years. That resistance would have given them an advantage over the plague-ravaged European farmers. So they moved in. With a lighter skin color and a proto-Indo-European language, this group and its migration still influence the look, languages and genes of modern Europe. According to Kristiansen, “it changed the course of European history. It changed the languages in Europe.” Genetically, he says, Europeans “are the descendants of those steppe people.”
Recently the research group found more evidence buttressing this theory of plague-driven change, when it detected Y. pestis DNA in two Swedish Neolithic skeletons dating from around five millennia ago. The disease, it appears, had arrived in Scandinavia just before the Yamnaya takeover. Kristiansen says his colleagues are now scouring “all over the place” for other instances of this early variant. He calls it “the mother of all plagues.”
An Evolutionary Journey
Several dozen ancient Y. pestis genomes now have been analyzed, from various points during the past 5,000 years. Changes in these sequences have enabled researchers to reconstruct an evolutionary history of the bacterium and indicate some early genetic alterations that may have helped transform an opportunistic gut pathogen into one of the biggest killers in human history.
In its earliest form—the 5,000-year-old variant—it was unlikely that the bacterium was carried by rat-riding fleas, as was the Black Death version. The older bacterium lacked an enzyme that the modern microbes use to prevent their digestion in flea guts. It probably spread through airborne droplets when its host—person or animal—coughed. But around 4,000 years ago Y. pestis gained a gene called ymt, possibly from another type of gut bacterium. (Bacteria swap genes frequently.) Ymt codes for that protective enzyme, which enabled the plague organism to live inside fleas and travel with the insects, says Johannes Krause, an ancient DNA specialist at the Max Planck Institute for the Science of Human History.
After acquiring ymt, the Y. pestis bacterium evolved the ability to form a biofilm—and it is this talent that is perhaps the microbe's most sinister innovation. Mutations appeared in a gene that improves the ability to produce an adhesive extracellular matrix, and other mutations hobbled different genes that would normally act to slow or stop matrix production. These changes enabled the bug to coalesce into sticky conglomerations of cells. These build up in the midgut of the flea, blocking its digestive tract. The starved fleas are pushed into a feeding rampage in which they repeatedly bite any mammal around, passing the bacterium with each nip.
People and Pathogens
The aDNA research has made it possible to trace the history of other microbes in addition to Y. pestis, enabling researchers to identify the dates when many modern human pathogens, including strains of leprosy, tuberculosis, hepatitis B virus and parvovirus, emerged as widespread troublemakers. Those dates, perhaps not surprisingly, occur when humans started to settle down, Krause says.
As civilization developed, distant communities were connected by horse, wheel, then boat—and wherever people went, the microbes went along with them. According to Herbig, long-distance trade facilitated “disease exchange on a global scale.” For example, the distribution of ancient hepatitis B and plague genomes follow well-documented human migration routes during the Bronze and Iron Ages. Likewise, tuberculosis was carried by “the crews of Roman trading vessels or the merchants who assembled at waypoints along the Silk Road,” according to Caitlin Pepperell of the University of Wisconsin–Madison, whose team used tuberculosis aDNA to estimate that modern strains emerged less than 6,000 years ago—rather than more than 70,000 years ago, as previously believed.
It was not just trade that spread these microbes. Pathogens often exploit multiple animal hosts, and the DNA data show that when our relationships with certain animals became closer, pathogens soon followed. One of the last remaining populations of red squirrels in the U.K., for example, still harbors a medieval strain of leprosy, possibly shipped into England by Viking fur traders. Similarly, a strain of tuberculosis that afflicts people was apparently transported to South America by seals, as revealed by aDNA from a millennia-old Peruvian skeleton. That tuberculosis genome is most closely related to strains found in modern seals and sea lions. “For this population, it makes quite some sense from an archaeological and anthropological view because they did a lot of seal hunting,” explains Herbig, who was involved with the research. “They created pottery on which you can find images of people seal hunting and also processing seal meat.”
This combination of human factors that increase vulnerability to pathogens—larger populations, greater global connectivity, an ever shifting relationship with the animal world—had a major impact on the New World when Europeans first arrived. The Aztec Empire, centered in Mexico, was invaded by a small contingent of Spanish forces in the early 1500s who toppled the civilization with the aid of disgruntled subjects and rival states. The Spanish then installed a brutal encomienda system of harsh treatment, overwork and malnourishment. And the European outsiders seem to have brought other attackers with them as well.
After the initial conquest in 1521, the Aztec population was devastated by one of the biggest pandemics in history. Written accounts from Spanish Franciscan friar Bernardino de Sahagún, who arrived in Mexico eight years after the initial Spanish contact, indicated that an infection killed off as much as 80 percent of the Indigenous population. But the identity of this cocoliztli pest (as the locals named it) remained a mystery. Guesses have ranged from hemorrhagic influenza to malaria to typhoid to smallpox. To historians, it was not even clear if the disease was of local origin or was imported by the Spanish.
In 2018, however, aDNA pointed to a likely culprit. Obtaining DNA from skeletons discovered in a cocoliztliera mass grave, Krause and his colleagues established that more than half of the samples had Salmonella paratyphi C., a bacterium that causes a severe intestinal disease. Salmonella organisms had not been found in the Americas before European contact, so it was almost certainly shipped across from the Old World. The conquistadors probably carried contaminated food and water on their transatlantic vessels, along with other potential vectors such as chickens, pigs, cattle, and vermin such as rats and mice. All were capable of transmitting disease.
At just this time an environmental misfortune in the Americas helped microbes such as Salmonella find a new home. A series of catastrophic droughts hit Mexico in the 1500s—established by tree-ring data published in 2000—and food shortages and population dislocation left people weak and unable to fight off unfamiliar microbial invaders that their immune systems were not prepared for. A civilization crumbled.
Today societies know much more about pathogens and how to fight them than did people 500 or 5,000 years ago. But our current struggles with COVID-19 show that our vulnerabilities to novel diseases have not changed: they often jump to humans from other species, spread via global trade and travel, and become exacerbated by crowding, poverty and malnourishment. The aDNA research reminds us of those enduring facts and shows that some of the biggest events in history were not just defined by powerful figures such as Emperor Justinian I or conquistador Hernán Cortés. They were also profoundly shaped by the microbes their empires helped to spread.