The 2019 novel coronavirus (2019-nCoV) behind the ongoing outbreak—which the World Health Organization has declared an international public health emergency—was named after the family of viruses it belongs to. The term “coronavirus” may have initially been unfamiliar to many, but most everyone has encountered milder forms of such viruses, of which four strains cause about a fifth of common cold cases. Other types cause diseases that are endemic in certain animal populations. But until less than two decades ago, all known human varieties caused illness so mild that coronavirus research was something of a backwater.
That all changed in 2003, when the pathogen behind the SARS (severe acute respiratory syndrome) outbreak in China was identified as a coronavirus. “Everybody in the field was shocked,” says microbiologist Susan Weiss of the University of Pennsylvania. “People started really caring about this group of viruses.” That outbreak is believed to have started when a coronavirus jumped from animals—most likely civet cats—to humans, resulting in a type of disease called a zoonosis. These viruses’ propensity for such jumps was underlined in 2012, when another virus jumped from camels to humans, causing MERS (Middle East respiratory syndrome). That illness has killed 858 people to date, primarily in Saudi Arabia, representing approximately 34 percent of those infected.
SARS, MERS and the new coronavirus almost certainly all originated in bats. The most recent analysis of the 2019-nCoV genome found it shares 96 percent of its RNA with a coronavirus previously identified in a specific bat species in China. “These viruses have been floating around in bats for a long time” without sickening the animals, says microbiologist Stanley Perlman of the University of Iowa. But there were no bats being sold at the animal market in Wuhan, China, where the current outbreak is thought to have begun, suggesting an intermediate host species was likely involved. This situation seems to be a common feature of these outbreaks. Such hosts may increase the viruses’ genetic diversity by facilitating more or different mutations.
But what is a coronavirus? What determines whether, when and how it jumps to humans and how infectious it will be? And what makes the difference between a case of the sniffles and a deadly disease? In the years since these viruses first emerged as a severe global health threat, researchers have been studying their molecular biology in an effort to answer such questions.
Anatomy of a Coronavirus
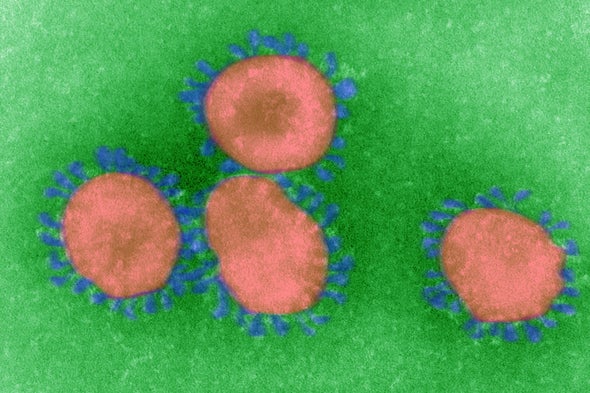
Coronaviruses are enveloped, single-stranded RNA viruses, which means that their genome consists of a strand of RNA (rather than DNA) and that each viral particle is wrapped in a protein “envelope.” Viruses all do basically the same thing: invade a cell and co-opt some of its components to make many copies of themselves, which then infect other cells. But RNA replication typically lacks the error-correction mechanisms cells employ when copying DNA, so RNA viruses make mistakes during replication. Coronaviruses have the longest genomes of any RNA virus—consisting of 30,000 letters, or bases—and the more material a pathogen copies, the more opportunity there is for mistakes. The upshot is that these viruses mutate very rapidly. Some of these mutations may confer new properties, such as the ability to infect new cell types—or even new species.
A coronavirus particle consists of four structural proteins: the nucleocapsid, envelope, membrane and spike. The nucleocapsid forms the genetic core, encapsulated in a ball formed by the envelope and membrane proteins. The spike protein forms club-shaped protrusions that stick out all over the ball, resembling a crown or the sun’s corona—hence the name. These protrusions bind to receptors on host cells, determining the cell types—and thus the range of species—that the virus can infect.
The major difference between coronaviruses that cause a cold and those that cause a severe illness is that the former primarily infect the upper respiratory tract (the nose and throat), whereas the latter thrive in the lower respiratory tract (the lungs) and can lead to pneumonia. The SARS virus binds to a receptor called ACE2, and MERS binds to one called DPP4—both are found in lung cells, among other places. Differences in the distribution of these receptors in tissues and organs may account for differences between the two diseases, such as the fact that MERS is deadlier than SARS and features more prominent gastrointestinal symptoms. MERS is not hugely infectious, however, which may also be a receptor-related trait. “DPP4 is expressed [highly] in the lower bronchi [airways leading into the lungs], so you have to have a large number of viruses coming in, because our airways are very good at filtering out pathogens,” says virologist Christine Tait-Burkard of the University of Edinburgh. “You need prolonged, intense exposure [to reach the lungs], which is why we see people who work closely with camels getting sick.”
Conversely, because pathogens can get in and out of the upper airways more readily, viruses that replicate there are more infectious. In addition, “the ability to replicate in different temperatures makes a big difference, because the upper respiratory tract is cooler,” Tait-Burkard says. “If the virus is more stable at those temperatures, it doesn’t go to the lower respiratory tract.” The lower airways are also a more biochemically and immunologically hostile environment, she adds. Analysis of 2019-nCoV strongly suggests the new virus, like SARS, uses ACE2 to gain entry to cells. This observation would fit with the fact that it appears, so far, to be less deadly than MERS (the current estimated mortality rate for the new coronavirus appears to be about 2 percent, but that figure may change as the outbreak unfolds and more cases are detected).
The picture quickly becomes complex, though, because viruses that use the same receptor can result in drastically different illnesses. One human coronavirus called NL63 binds to the same receptor as SARS but only causes upper respiratory infections, whereas SARS primarily infects the lower respiratory tract. “Why that is, we don’t know,” Perlman says. Another curiosity is that the ACE2 receptor is prevalent in the heart, but SARS does not infect heart cells. “That was a clear indication that other receptors, or co-receptors, are also involved,” says molecular biologist Burtram Fielding of the University of the Western Cape in South Africa. The virus binding to a receptor is only the first step in the cell entry process. When a virus binds to a host cell, they start morphing together, and other viral proteins may bind to other receptors. “For the efficiency of entry, it’s not just the one main receptor,” Fielding says. “There could be others as well.”
Immune-System Arms Race
Another important feature of coronaviruses is their “accessory” proteins, which appear to be involved in evading the host’s innate immune response—the body’s front line of defense. The response is initiated when a cell detects an invader and releases proteins called interferons, which interfere with the pathogen’s replication. The interferons trigger cascades of antiviral activity, from shutting down host protein synthesis to inducing cell death. Unfortunately, most of these processes are also bad for the host. “A lot of the disease that’s caused is actually the immune reaction—inflammation—and destructive things induced by viruses,” Weiss says. “That will also determine how virulent a virus is: How much of a destructive immune response does it induce, as opposed to just a protective one?” This aspect is also why underlying medical conditions are so important. Most of the people who have died from the new coronavirus to date “had comorbidities, like autoimmune diseases, or secondary infections, which can become much more prevalent once our innate immune systems are busy fighting a virus,” Tait-Burkard says. “That’s why the important thing is to treat people for comorbidities and give them antibiotics to stop bacterial infections taking hold.”
Of course, the immune response’s purpose is to eliminate invaders, so viruses possess countermeasures. This trait seems to be what differs most among various coronaviruses. “These viruses are closely related, but they have different accessory proteins,” Weiss says, adding that they “have evolved to shut down various aspects of that [innate immune] response.” Some researchers think bats harbor coronaviruses because they do not mount the intense immune response humans do. “A lot of the signaling molecules that alert our immune system are suppressed in bats, so they don’t get sick,” Tait-Burkard says. Rather than reacting, bats maintain a constant low-level response, which may contribute to the viruses’ evolution. “[Bats] have a constant expression of interferons, which selects for viruses that are good at evading that response,” Tait-Burkard says. “So bats are very good selection vessels for viruses that are very good at hiding.”
Accessory proteins are far from fully understood, however. “They can be taken out of some viruses without any effect on the ability of the virus to grow,” Perlman says. “You would think: if you had a protein that was key for countering the immune response, if you took it out, the immune response would win—and it’s not necessarily so.” Some researchers believe accessory proteins influence how deadly coronaviruses are. There have been studies with SARS in which removing an accessory protein did not change the virus’s replication efficiency, but it became less pathogenic. “Lots of virus would still be made, but it seemed to be less harmful,” Fielding says.
Coronaviruses do possess some ability to correct genetic errors, but it neglects certain regions of their genome, Tait-Burkard says. Consequently, two sections, in particular, are especially prone to mutations: those that encode the spike protein and accessory protein regions. “In those two areas, coronaviruses allow a lot of mistakes, which drives their evolution, because they manage to bind to new receptors and evade the immune response of new systems,” Tait-Burkard says, “which is why coronaviruses are so good at jumping from species to species.”