Brain cancer is terrifying. It attacks an organ we see as the core of our personality, our mind, our very humanity. And because the disease grows inside the brain, it is notoriously difficult to treat. The organ has evolved many defenses to keep foreign substances out as a method of self-protection, but those substances include many anticancer drugs. Using knives or radiation on this citadel of consciousness carries tremendous risks. For these reasons, the five-year relative survival rate for people aged 55 to 64 who get glioblastoma, the most common type of primary brain tumor, is a grim 5 percent. The disease killed John McCain, Edward Kennedy and Beau Biden, and it takes the lives of about 15,000 less famous Americans every year.
Now we have developed a nano-sized drug that travels through the body and into the brain, where it can kill off cancerous cells. These drug particles are composed of oligonucleotides—strands of DNA or RNA, the molecules that make up the master code that tells every cell what to do—and they stick out from a central core like the many spines of a sea urchin. The spiny round particles are called spherical nucleic acids. In an early trial with eight patients, these spheres went into glioblastoma cells and bound up other “code” molecules that are key to the cancer’s incessant growth.
Such spherical drugs appear to work against a variety of diseases. Another terrible affliction, this one affecting infants, is spinal muscular atrophy, or SMA. It robs children of muscle control until swallowing and breathing become first difficult and ultimately impossible. Most youngsters with the disorder succumb before they enter kindergarten, and until recently there was no help doctors could offer. In 2016 the U.S. Food and Drug Administration approved one remedy: a drug called Spinraza that is injected directly into the spinal cord several times every year and, at a list price of $125,000 per shot, is one of the most expensive drugs in the world. We recently compared our spheres, studded with nucleic acids that get inside cells and interfere with messenger molecules that lead to SMA’s symptoms, with the Spinraza approach in studies of rodents. The spheres improved survival by four times—115 days versus 28 days—and the rate of toxic side effects was much lower.
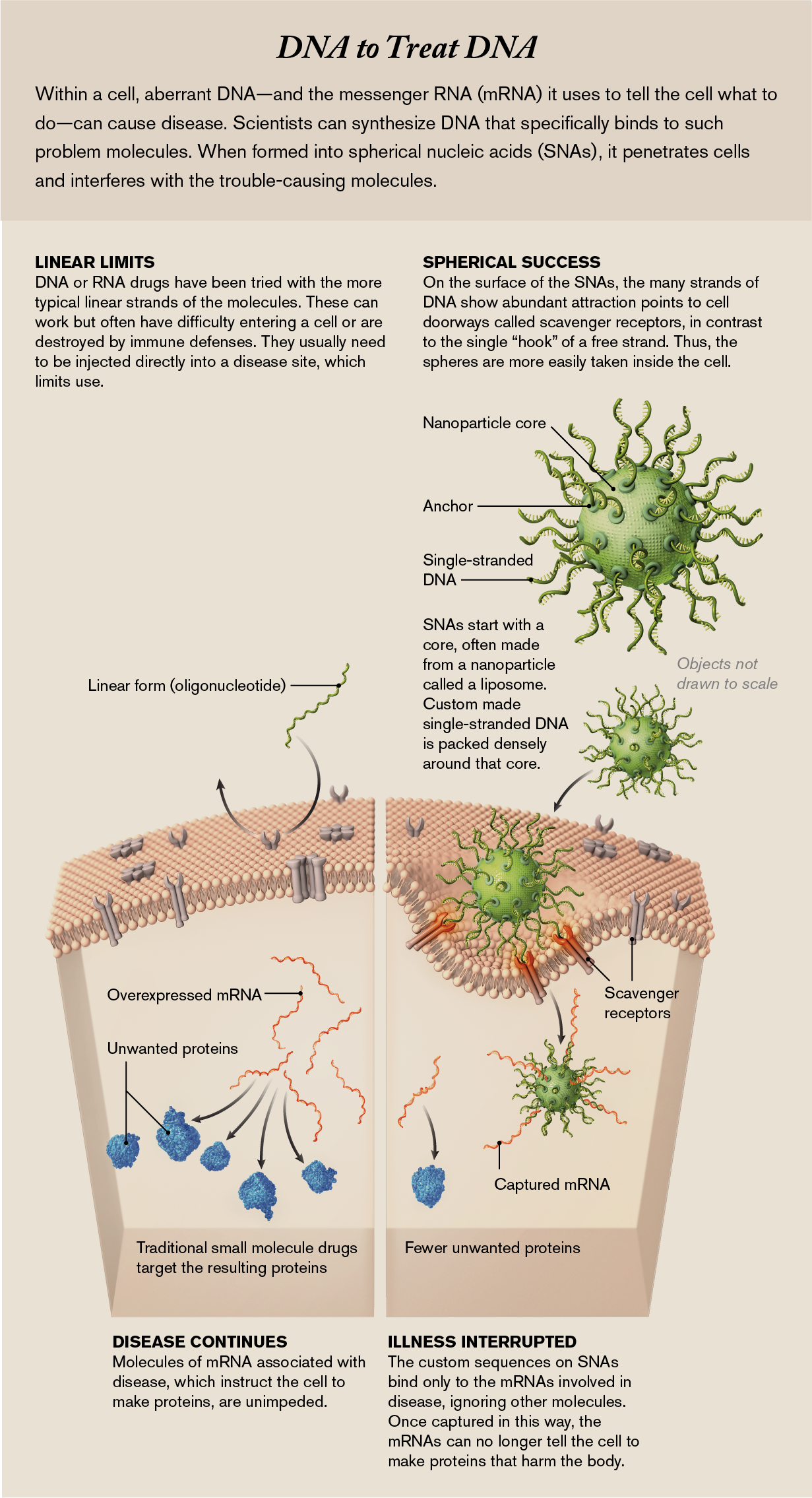
Spherical nucleic acids, or SNAs, avoid problems that have plagued the pharmaceutical industry’s attempts to develop new drugs. Conventional drugs are nonspecific: they can affect many cells and organs, not just diseased ones; hence, they have numerous side effects. Nucleic acids, however, can be designed to interfere with only disease-causing genes or their related instruction molecules sent to control a cell’s behavior. Biologists have tried to use nucleic acids in the past but primarily as linear molecules and with little ability to direct where they go. And because the body has robust defenses against foreign genetic material—the immune system, for one—in most cases, these defenses damaged the drugs immediately or sent them to organs such as the liver and kidneys for waste removal.
But SNAs, at only billionths of a meter across, seem able to travel anywhere in the body and get inside cells before immune defenses can waylay them. The spherical shape lets us pack a high density of nucleic acid “spines” into a small space, and that density creates a strong interaction with receptors on cell surfaces that admit the particles inside. There the sequence of the components—the same nucleotides, abbreviated as A, T, C and G, that constitute the DNA code of life—ensures that they affect only complementary sequences of DNA or RNA. (The latter molecule uses U—uracil—instead of T, and we design for that.) We construct our strands to match only sequences in the cells that are crucial to the disease. SNAs are not magic bullets and will have to pass many more tests before they can be used on lots of patients. But the potential is there: because the nucleic components can be reordered to interfere with many different disease-causing molecules within cells, the spheres have the ability to tackle some of the world’s most debilitating conditions.
Programmable Drugs
Traditionally, scientists have found disease treatments by screening hundreds of thousands of small synthetic or natural molecules, going through a long trial-and-error process to see if any of them have therapeutic benefits. Although this pipeline has led to a number of amazing medicines, such as antibiotics, even the most promising ones can cause unwanted side effects. Many other diseases are unaffected by these molecules and therefore still lack a cure or treatment. Even biologics, a newer class of drugs that are often based on proteins made by immune cells of mice, rabbits and other animals, typically rely on an abbreviated trial-and-error discovery process.
An ideal drug-design process would allow scientists to rapidly and rationally design specific drugs that use the same language as our cells, instead of looking for a needle-in-a-haystack molecule. Cells communicate many complex messages through DNA and RNA to make millions of proteins. The number of steps that cells must execute correctly to make these proteins is staggering: they must select a specific sequence of DNA made of A, T, C and G nucleotides, transcribe that sequence into a form called messenger RNA (mRNA), and then accurately read that mRNA to arrange molecules called amino acids into a chain—as long as 35,000 units—that forms a single protein.
Errors where one nucleotide such as a T or a G is added, deleted or placed in an incorrect order can halt protein production or generate an irregular protein that causes disease. Too many copies of an mRNA, and therefore of its related protein, can also lead to disease. (So can the introduction of foreign nucleic acids from a virus, which leads the infected cell to make harmful viral protein.)
But we can synthesize our own stretches of DNA or RNA components, called oligonucleotides. Because the genetic alphabet has very specific rules—A can bind only to T, and C binds only to G—we can make our oligonucleotides with sequences that selectively bind to and inactivate one disease-driving sequence. When they do so, the synthetic oligonucleotides gum up the cellular works, preventing the affected cells from producing a disease-causing protein.
Yet despite automated equipment that can rapidly make synthetic oligonucleotides with any desired sequence one could imagine, fewer than a dozen oligonucleotide-based drugs have been approved for patients. This is because these strands of oligonucleotides face a significant hurdle once they are injected into the bloodstream: because they are foreign—that is, not native to the patient—they get treated as hazardous material or waste. The body’s immune system either destroys these oligonucleotides, or the body’s waste-filtration stations, the liver and kidneys, remove them. They do not reach their intended target. Even if oligonucleotide strands could make it to a cell that contained the target mRNA, that cell has an outer membrane that acts as a barrier to prevent the oligonucleotides from getting inside. As a result, drug companies working with oligonucleotides have often settled for treating diseases that can be targeted in the liver. The liver is an important organ. But sequestering these drugs in this one place really limits their use. (An alternative approach—injecting oligonucleotides directly into the disease site, such as into the spinal column with Spinraza—is technically difficult and still does not ensure entry of the medicine into all the appropriate cells.)
A Surprising Result
Advances in nanotechnology made by our group at Northwestern University, along with several other researchers, have led us to the SNAs, which may be a way around this problem. Prior to 2006, our group had been interested in using the highly specific binding ability of SNAs in probes for ultrasensitive diagnostics—to fish out stretches of cancer DNA from blood samples, for instance. We could do this by chemically decorating a gold nanoparticle with many strands of DNA designed to anchor one end to the particle, producing the sea urchin spine pattern. The outer end of the DNA was designed to be a complementary sequence to the cancer DNA sequence, so it worked nicely as a probe. We also used the spheres as artificial atoms with programmable bonds to fashion new types of materials. Drug design, however, was not really on our radar. After all, according to the dominant paradigm of drug biology and chemistry, RNA and DNA would not naturally cross cell membranes.
We were curious, though, about how nucleic acids in this new geometry would interact with living systems. Drug developers had already been experimenting with single strands of oligonucleotides, with, as we noted, limited success. From our research with SNAs as a diagnostic platform, we knew that target DNA and RNA would bind to our clusters of spines much more strongly than they would attach to free oligonucleotide strands. The reason is that our spines are packed densely on the nanoparticle’s surface. That makes them more rigid, which helps the As, Ts, Gs and Cs on each strand align and bind when they encounter a target strand. This characteristic made us suspect that with the right nucleic acid sequences, SNAs could be a very potent oligonucleotide drug.
To test this idea, we carried out an experiment that, at the time, we thought had only a slim chance of working. We took strands of free oligonucleotides and put them into a test tube with mouse cells. In a different tube we added a bunch of SNAs to the same type of mouse cells. We attached red fluorescent molecules to both the spheres and the strands to help us track them. When we looked at the cells under a microscope, the ones mixed with free strands appeared transparent, as expected. Free oligonucleotides did not cross the cell membrane. But the cells mixed with SNAs lit up the screen with bright red fluorescence. The spheres had made it inside!
How could this happen? In general, cell membranes closely regulate which molecules may enter, and oligonucleotides are not typically among the approved guests. Furthermore, oligonucleotides carry a negative electrical charge, as do cell surfaces. Like two magnets, the two biological objects should repel each other. Yet when we repeated this experiment over and over again using more than 50 other human and animal cell types, all but one glowed red, a signal of success.
Today we think we know what the gateway is: a type of doorway molecule called a scavenger receptor that dots the cell surface. These receptors play a major role when a cell engages with its environment; for example, they admit nano-sized biomolecules the cell needs. Some of the structural features at the ends of SNA spines happen to mimic the natural substrates of these scavenger receptors. As noted earlier, the strands on the spheres are densely packed, and like with Velcro, the more hooks, the stronger the bond. With free strands, even if scavenger receptors recognized them as molecules to take in, they have only one hook and float away.
With the aid of an electron microscope, we could see that once an SNA binds to these receptors, the surrounding cell membrane folds inward to create a pocket, ushering the SNA into the cell.
Spheres as Medicine
But getting in was only half the battle. To work as a drug, the SNA needed to find, bind to and inactivate a particular stretch of mRNA that instructed the cell to make a disease-associated protein.
The first stretch of mRNA in a cell that we targeted did not cause disease but did instruct the cell to make a protein that glowed bright green under a microscope. Our goal was to stop this mRNA. When we exposed mouse cells to an SNA designed to match that green-causing mRNA and compared them with similar cells that did not get the spheres, the color difference was clear. Sphere-free cells were bright green, showing the mRNA had encoded proteins. But cells exposed to our SNAs were transparent, meaning we had blocked the mRNA before it could pass along instructions to make anything green, as we reported in Science in 2006.
Next we pitted SNAs against the major challenge plaguing linear oligonucleotide drugs: destruction by the body’s natural defense system. We found that our spheres have a strong electrical charge—again because of the dense packing—that helped them evade immune interference. This high charge inhibits defense molecules called nucleases, proteins that degrade foreign DNA and RNA, from getting close.
Reality Test
We were on to something, at least in the laboratory. Other scientists replicated and independently advanced some of our work, including dermatologist Amy Paller, Arthur Burghes, an expert on SMA, immunotherapy specialist Bin Zhang, cancer biologist Alex Stegh, transplant surgeon Jason Wertheim, and oncologist Priya Kumthekar. But the path from benchtop breakthroughs to healthier patients is long and hard, so nearly 10 years ago researchers from our group founded a company called Exicure to advance SNA-based drugs to the clinic.
We initially explored whether these potent drugs could be delivered to diseased tissues in skin creams and eye drops, which is feasible because SNAs are easily taken up by cells and a big improvement over invasive strategies such as direct injections. Two of our first targets were psoriasis and poorly healing wounds, and there are several promising SNA candidates already in early-stage clinical trials for some of these ailments.
Skin, of course, is relatively easy to get to. The brain is not. Defended by a vigilant immune system and a web of blood vessels—the blood-brain barrier—designed to keep foreign molecules out, the brain makes cancers such as glioblastoma particularly difficult to treat. We thought, however, that SNAs might move across these defenses via the same doorway molecules that ease their path through cell membranes. Once in the brain the spheres could home in on cancer cells by targeting genes and proteins responsible for keeping the cells alive, which malignancies produce in excessive amounts.
To start this project, we created an SNA drug with many short pieces of RNA specifically designed to knock down the production of a protein in glioblastoma cells called Bcl2L12. That protein acts as a biochemical defender that helps to keep the cancer cells functioning. We thought that by intercepting the mRNA that tells the cells to make this protein, the SNAs could make the cancer vulnerable to conventional medicines. Indeed, in our animal studies, reported in 2013 in Science Translational Medicine, that is what happened: SNAs injected into the bloodstream of mice reached the brain, crossed the blood-brain barrier and prevented the production of Bcl2L12 protein inside of glioblastoma cells. Last year early clinical results showed that these SNAs also reach glioblastoma cells in human patients. We did not cure people, and we have yet to test whether the SNAs make the cancer cells more vulnerable. Still, the ability of SNAs to reach the brain and their lack of toxicity generate hope for treating this cancer, as well as other neurological disorders, and set the stage for the next set of clinical trials. And tests in other diseases, such as spinal muscular atrophy, show promise in animals.
Another exciting direction for SNAs is their use as immunotherapies against cancer. Cancer cells often have proteins in their membrane that are different from the proteins found in healthy cells. Therefore, a cancer cell protein can act as a red flag, and if our immune system can be trained to go after it the way it goes after a flu virus, our own bodies can do a better job of protecting us from the disease.
To make an SNA cancer vaccine, we exchanged the gold-nanoparticle core for a hollow nanoparticle called a liposome, filled it with one of these red-flag proteins and injected it into animals with the corresponding cancer. Some of our most recent experiments, published in 2019 in the Proceedings of the National Academy of Sciences USA, showed that such SNAs elicit an immediate immune response to the tumor, apparently teaching the immune system to go after cells showing that red flag. The effects appear long-lasting, too: the immune system keeps going after cells with that protein after the SNAs have vanished. SNAs are already showing potency and safety in phase I clinical trials in humans, and other spheres targeting a deadly skin cancer are being tested in a separate set of safety trials.
SNAs are, however, not yet approved drugs. There are a number of challenges that they have to overcome first. Because the spheres do get to a wide set of cells, we need to carefully study whether or not they produce any negative “off target” effects even though their design should limit them to only problem DNA and RNA. Larger patient populations must be explored, and we need to improve targeting to increase the amount of drug that gets to the affected organ and cells.
We think the ability of SNAs to access so many different tissues is game-changing and will be central to the emergence and ultimate widespread use of such medicines. SNAs are the product of three core capabilities: the ability to make large quantities of oligonucleotides, an understanding of genetic disease pathways, and the ability to get such oligonucleotides into tissues and cells that matter. The first two are important, but without the third the process is like making software without hardware it needs to run on. SNAs may be that crucial and versatile hardware—a platform able to be reused for many different types of illness, one that begins to move the pharma industry away from the difficult search for entirely new molecules for every new treatment. An SNA simply needs a different set of oligonucleotides to be sent after a new disease. And we are just getting started.